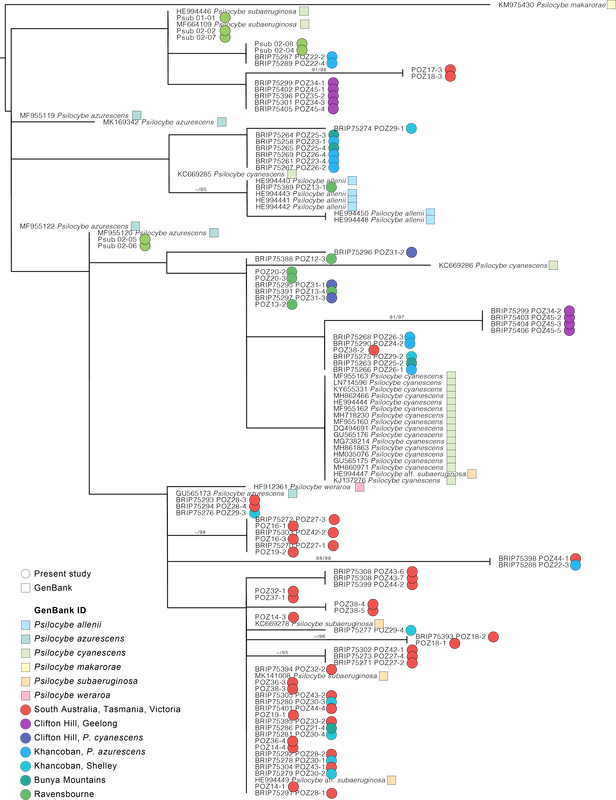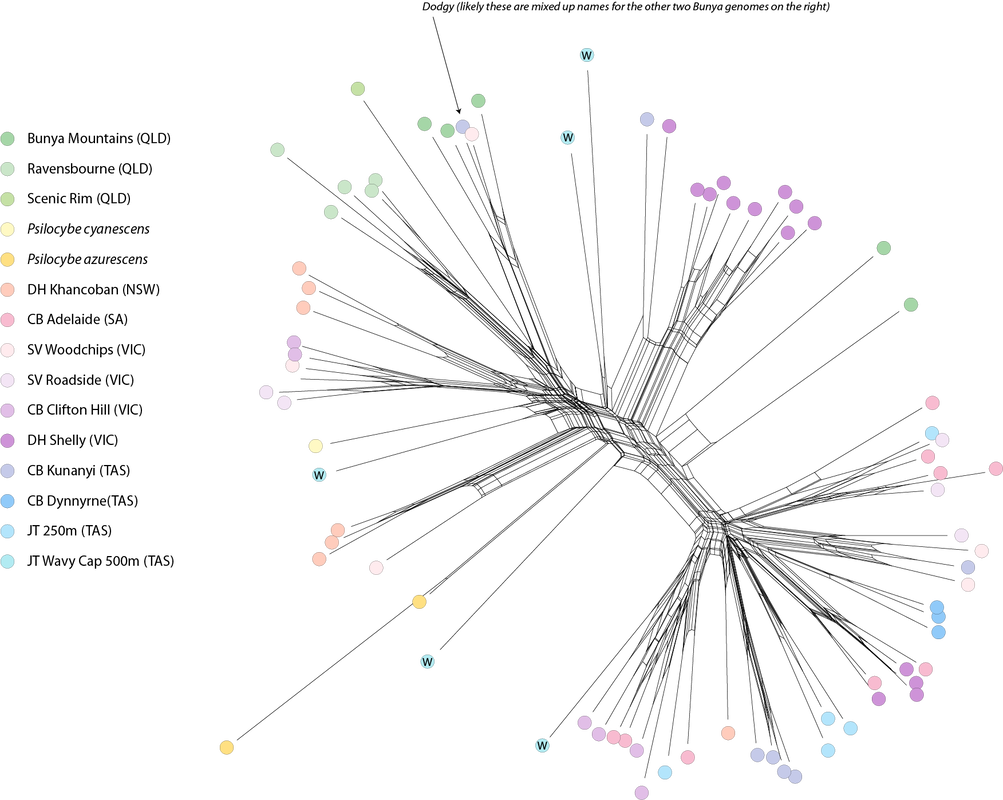|
Taxonomy is the bedrock of communication about life. If other facets of communication changed, it'd be pretty frustrating, e.g., "from here on, hello will mean mustard". I witnessed the saltiness of taxonomic change first hand in South Africa. Africans had spent their lives calling their iconic, thorny trees Acacia. Acacia even means thorny. That didn't suit taxonomists and all the African acacia were renamed to Vachellia. The non-thorny Aussie trees, of which there were >1,000 species compared to 100-ish in Africa, took the name. Ja, I can see why they'd be salty. In other areas of science, we support or reject hypotheses and move on. Taxonomy is descriptive, hypotheses can be loosely applied (we hypothesise this is a monophyletic group of taxa), but it's a stretch. When two people describe the same piece of art or music, they may differ in their opinion. The same is true for taxonomy. Applying taxonomic ranks to organisms is arbitrary and cases can be made to split or lump any rank at a shared common ancestor. Ultimately, determining gene flow among populations paints a clear picture of species boundaries, but who has the time to do this for millions of fungi. I find it interesting that we become attached to taxonomic names, even when there is evidence to support that they should be called something else. I think we are more emotional because of the vagueness in species/taxon delimitation. In the smut and rust section of this blog, you can read about the taxonomic name for corn smut and why it should be called something other than Ustilago (but jeeze, you'd have to be bored). Clearly even I become emotional about taxonomy. Taxonomic names, as long as they were validly described, are always there for communication and can be used eternally (as long as people know what organism you mean). Just 'cause some fella is saying all the northern hemisphere wood-loving shrooms are Psilocybe subaeruginosa doesn't mean you have to forgo these names. Keep on using them... what do you think African people call acacia (hint: not Vachellia)? The only change here is that we know Australia is the centre of origin of P. subaeruginosa and it has spread to the northern hemisphere where multiple new names have been applied to one taxon. 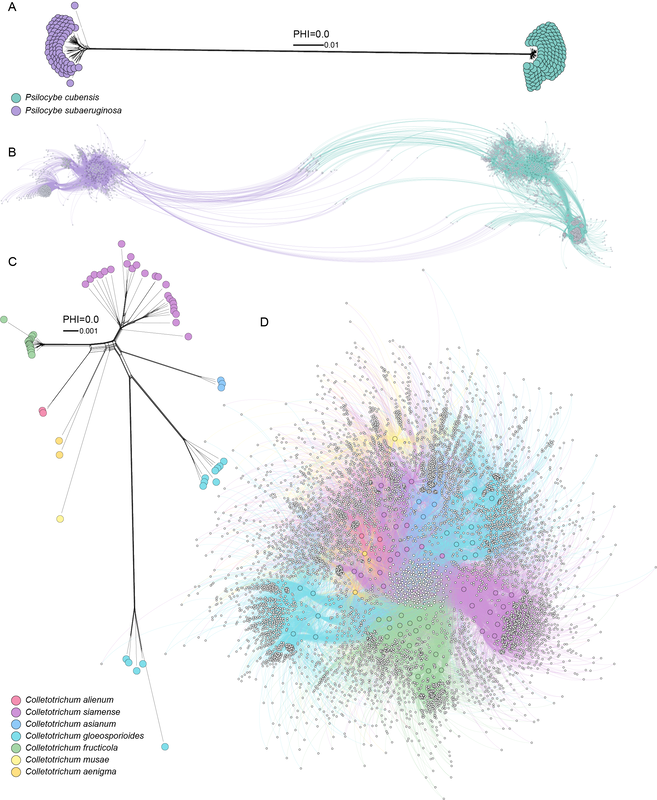 Population diversity illustrated for two species of Psilocybe and several species of Colletotrichum. A. SplitsTree neighbor network based on 382 single copy orthologs identified by OrthoFinder between P. cubensis and P. subaeruginosa. Edge length reflects genetic difference and reticulation is an indication of recombination. B. Network of relatedness among genomes (large circles) and clusters of accessory orthogroups (small grey circles), with nodes and edges coloured by species of Psilocybe. C. SplitsTree neighbor network based on 3,144 single copy orthologs in the Colletotrichum gloeosporioides species complex. The scale is a magnitude smaller than between species of Psilocybe. Long edges radiating from reticulation is a signature of clonal reproduction. A test for the pairwise homoplasy index was 0.0, which indicates randomness of alleles across the alignment and is evidence of reproduction. D. Network of relatedness among genomes (large circles) and 7,989 clusters of accessory orthogroups (small grey circles), with nodes and edges coloured by species of Colletotrichum. Species cluster together to some extent based on their shared accessory genes, however, there is no strong separation of genomes and accessory orthogroups are shared among taxa. The figure illustrates genomic relationships based on core and accessory genes between sister species. For magic mushrooms, the sisters are cubes and subs. Note they are highly separated by core genes and there is almost no overlap of accessory (non-core) genes. Colletotrichum is the other example, and it has been split like a banana in a dessert bar. Note that the core and accessory don't clearly separate species. Likely these are all the same taxon and taxonomists have described clones as different species (which could be acceptable in some instances).
After this explanation, if you still prefer species complexes and lots of names for the same taxon, why not spend your time and resources examining the boundaries of recombination to demonstrate speciation in process?
1 Comment
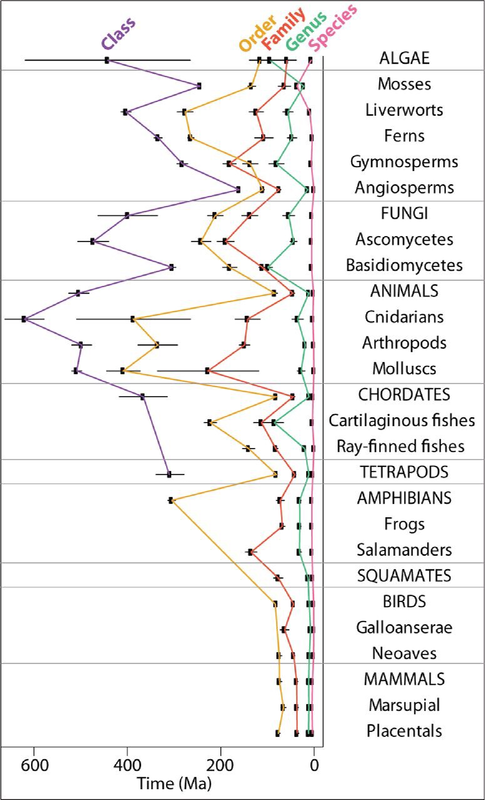
Set and setting, and psilocin binding to 5HT2A receptors are the two domgas of how magic mushrooms work. To the point it feels that's all we think matters to dictate a psilocybin experience from magic mushrooms. Let's think back a couple of million years to before mammals were on the scene. Under a clock-like rate of speciation, species are between 500K to 5 million years old. Genera are somewhat older, and Basidiomycota are an anomaly compared to other organisms. Genera of Basidiomycota are usually 20–100 million years old. (Check out this paper if you want to read more: https://academic.oup.com/mbe/article/32/4/835/1078218). Psilocybe could be anywhere from 20 to 100 million years old. We assume that the most recent common ancestor of all species of Psilocybe could produce psilocybin (because all except one produce it), and likely psilocybin is the same age as Psilocybe. All species of Psilocybe, extant and extinct, have forged their way in niches of leaf litter, wood, grass, moss, and dung. We don't know the exact purpose of psilocybin, whether to repel, control, or attract, but we can be certain it targets metazoans with serotonin receptors. Whatever metazoans dominate the different niches of Psilocybe, we can be sure over an evolutionary time scale the genes that produce psilocybin in 200 or so species are refined to best target species of slug, arthropod, nematode, or whatever was eating it for whatever reason in whatever niche. This has happened time and again with co-evolved relationships, especially those with metabolites governing the symbiosis. If you accept that there are different predators across 200 species of Psilocybe, you may be on the way to understanding why I think different magic mushrooms give different psychedelic experiences. Different effects from different species given an evolutionary time scale is hopefully palatable, but to say that allelic variation in one species changes an experience is more difficult to swallow, especially with the dogmas of how trips work. We showed that populations of P. subaeruginosa maintain genetic diversity in the alleles that produce psilocybin rather than just one becoming dominant (balancing selection to be fancy). This is probably why WLP can be in some, but not all genotypes and is maintained in populations. What the blazes is the benefit of all this diversity? Diversity to the extent that psilocybin alleles are barely shared by closely related populations and there is reason to suspect recombination within the locus itself that would ensure that the same alleles in the pathway are not linked (or not always inherited together).
Long story short, our work on subs shows an evolutionary process of maintaining allelic diversity in the psilocybin pathway (or balancing selection). Cubes have a solid 5–6 alleles at the psilocybin locus in the entire population of cultivated mushrooms and a bunch more in naturalised populations. I've grown most of these and am in the process of testing the tryptamine landscape in homozygous genotypes. I've crossed different alleles to see what happens when the psilocybin locus is heterozygous. I've never been more convinced that there are phenotypic impacts from genotypic diversity, and I hope to hold hard evidence soon. Rest assured, if the time comes, the people will be able to decide if there are differences themselves. In the meantime, do me a favour next sub season: don't combine harvests from different patches. Experience them separately and share whether there is a difference (given a consistent set and setting). Expect there to be different psilocybin alleles at different mushroom sites, unless the mushrooms are harvested from mulch/woodchips that have been spread artificially (wild harvests will guaranteed be different alleles). More on all this soon. Full piece at Current Biology here:
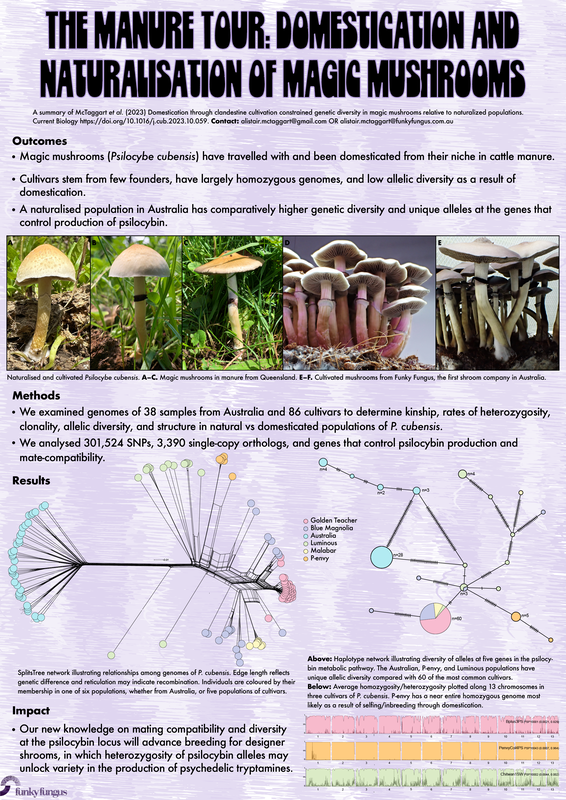
https://www.sciencedirect.com/science/article/pii/S0960982223014604?dgcid=author. And just in case you hadn't seen that our sub work is publicly available, just not peer reviewed right here: https://www.authorea.com/users/700719/articles/687526-wood-loving-magic-mushrooms-from-australia-are-saprotrophic-invaders-in-the-northern-hemisphere. A hint to the end of my research career came the first time I met the Institute Director at QAAFI in November '22. He opened our online meeting with 'Hello, your work doesn't really fit in here does it?' Bare-chested, riding the unicorn of the TGA announcement in February '23, Chris Appleyard, the first person to legally grow magic mushrooms in Australia (to my knowledge), called me up and offerred me a job. A stroke of good fortune for yours truly. My role at Funky Fungus is in designer shrooms and we have approval to fruit mushrooms. This was something missing from my permission to work on shrooms at UQ. Chris has stocked the Funky Fungus library with all of the cultivars I've researched, and what a pleasure to finally get to see them in the flesh. Chris owned one of the properties where we first collected Psilocybe cubensis back when I began studying magic mushrooms (the Woodford specimens, specifically Delaney's Creek, if you want to see where they fit in the Australian population in the Manure Tour or posts below). Where's all this going? I'll keep the blog going because we have a research grant to characterise the concentration of psilocybin (and related tryptamines) of 100 genotypes in the FF library and to compare differences between growth treatments (such as blue light while pinning, degradation of psilocybin after harvest, mycelium vs mushroom, and whatever else we think of). I also thought I should explain the reasoning behind designer shrooms and why we're crossing different haplotypes. Here's a brief outline of the process: fruit cultivars, make haploid cultures, cross against Oz haplotypes, identify cool phenotypes and genotypes that produce high or different concentrations of tryptamines. Who knew I'd get to put all the knowledge we've found to good use! 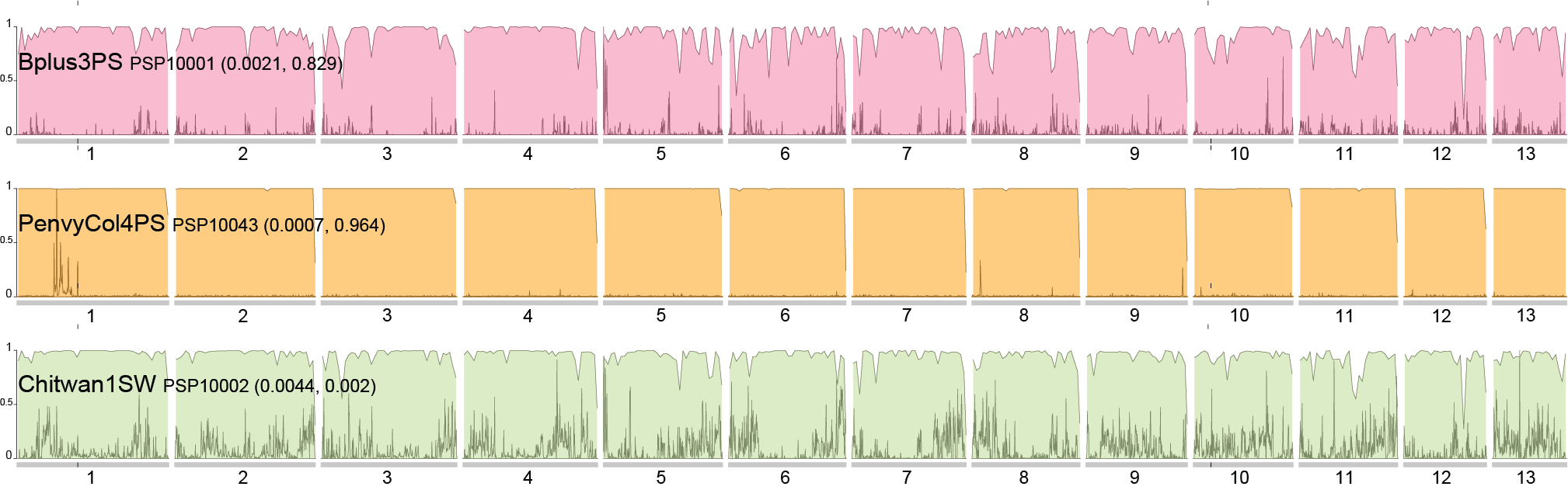 This figure illustrates heterozygosity and homozygosity across a genome. The density of homozygous (the big blocks of colour) and heterozygous (the sharp thin peaks) positions is plotted in 100,000 and 10,000 base pair windows across 13 chromosomes of Psilocybe cubensis. The genome of Penis Envy is incredibly homozygous, B+ a tad more heterozygous, and the Chitwan cultivar the most heterozygous of everything! You've heard me harp on about Penis Envy for ages. It's one of the most interesting cultivars to me because of very different alleles at the psilocybin locus compared to most other cubes (Treasure Coast is the other interesting cultivar), and P-envy is highly homozygous and will be pretty cool to cross against. The images below are some of the current crosses we've made over the last two months by getting haploids of a few cultivars (Amazonian, Costa Rica, Mazatapec, and Mexicana), and crossing them with a couple of Oz haplotypes. Really cool to see the different Oz haplotypes totally change the appearance of the mushroom. Now, we just need to determine how much of a bang they have for psilocybin and start crossing against Penis Envy and Treasure Coast, with an aim to have heterozygous alleles for psilocybin production and see how it effects the finished product.  Just over two years ago I started this research journey on magic mushrooms. I feel privileged for this opportunity and to have joined a community fascinated by these Fungi. Some new knowledge may have slipped by over two years, and now, at the end, it seems like a good time to summarise the outcomes, knowledge gaps, rejected hypotheses, and my predictions. Psilocybe cubensis Is naturalised in Australia. We know this because there is a large drop in its effective population size, followed by a recovery. It has very low mitochondrial diversity (we expect high diversity in a centre of origin), low diversity at psilocybin loci, but reasonably high diversity at mating loci because it has been outbreeding since its arrival. Psilocybe cubensis probably does not infiltrate the soil, which is why it doesn't occur in soil sequencing data. Rather it colonises manure, develops its hymenium, fruits, spreads new spores that are probably eaten by livestock, and then they come out the other end. Each genotype is ephemeral, which is why temporal sampling over three years at Tallebudgera shows unrelated genotypes. I believe there is population structure by geography, but movement of cattle and manure helps long-distance dispersal. Cubes were certainly not introduced intentionally by recreational growers. They've been here too long and there is no shared ancestry with cultivated lineages based on genetic analyses. There is more allelic diversity of P. cubensis in one piece of manure in Australia than in entire cultivated lineages. With this reservoir of diversity, someone with the right laminar flow could fix traits unseen by the community and put some 'spice' back into cultivated cubes. The majority of cultivated P. cubensis either came from one naturalised population or one genotype, it's hard to tell (to understand this, look at some of my cube networks with knowledge that clusters in Australia all came from one parental genotype). Cultivars like Penis envy have been completely inbred to the point where there is almost no heterozygosity in their genomes. This is part of the breeding process to fix traits over time, and may explain why 'a cube is a cube' despite unexplored diversity waiting for innovation.  Psilocybe subaeruginosa Has a centre of origin in Australia and is surely an example of a saprotrophic invader in the northern hemisphere. We have high phenotypic diversity in undisturbed national parks, high diversity at mitochondria, mating genes, psilocybin loci, and even in ribosomal DNA (like the ITS region). Whether the diversity of P. subaeruginosa translates to taxonomic diversity is tricky. Mushrooms must differ at their mating loci to cross, and separated populations may maintain connectivity through mate compatibility even after long periods of separation (as long as they still recognise each other). If it were up to me, I would treat everything as one phenotypically diverse species, P. subaeruginosa, which has limited gene flow among some spatially separated populations. Subs are perennial. That patch you visited in the 90s is the same mycelium you visit now. That mycelium will spread outward, as that genotype moves in its little area, fights other genotypes and decomposes wood and leaf litter (thank you subs). This conclusion is based on what you there in the community tell me, and the prolific sibling relationships recovered from many sampled mushrooms. For example, we recovered sibling relationships from many sampled mushroom pilei whether adjacent to each other, or further apart (such as Geelong and Clifton Hill). There is a lot of diversity in the psilocybin locus, and one mushroom can be heterozygous for different alleles in the pathway. Jeeze those three copies of psiH must do something! I favour a hypothesis that wood lover's paralysis is caused by a tryptamine analogue metabolised by one of these 'rogue' psiH genes and that binds to peripheral serotonin receptors. Have you seen the curative effects of MDMA on patients with Parkinson's Disease (check them out)? Before I tap out of shroom research, I'll make sure to share the cultures of suspected wood lovers (based on psiH) with people smarter than me. Entourage effect
I am a geneticist and believe that genetic differences translate to phenotypic differences. There is allelic diversity in the genes that metabolise tryptophan into psilocybin in both cubes and subs. I think this is why people have different experiences from different patches of mushrooms. When I read studies that conclude there is no difference in the experiences of psilocybin, LSD, and mescaline (like this one) my mind boggles. Is this because they use synthetic psilocybin? Is it because we don't have the language to describe these different experiences? I wish I knew. I look forward to meaningful research on the entourage effect, and whether this can be linked to genotype at the psilocybin locus (which controls the phenotypic concentrations of active tryptamines). Maybe designer shrooms will be a thing in the future. Diversity of hallucinogenic fungi in Australia The metabolic diversity of fungi in Australia is woefully understudied. Somewhere up in the NT is a magic mushroom in our soils, only a matter of time for its discovery. Based on the BASE soil sequencing data, there are only four species of Psilocybe in Australian soils. The dung niche must have more. If P. cubensis does not show up in soil sequencing data, maybe other dung inhabiting fungi are the same, and more await discovery. I look forward to seeing the discoveries of hallucinogenic mushrooms that must be present in our tropical rainforests (just as there is rich diversity in PNG). If you're on the ground, get spore prints, these just make life easier for the next mycologist :). The future of psilocybin use in Australia What an exciting time where we get to watch a stigma dissolve. But there are hurdles yet. Magic mushrooms have been used safely for millenia, however we hear that more time is needed to ensure their safe use. What makes them unsafe? (i) temporary psychoses (=a bad trip), (ii) impaired judgement during an experience, (iii) counter-indications with other medications like SSRIs, and (iv) legal penalties for their possession. We've pinned our hopes for psilocybin on the medical community who have an interest to control the use of psilocybin (at $25,000 for treatment is the recent estimate I read). I'm reminded of the adage 'never ask a barber if you need a haircut.' The dogma of clinical trials, which rely on medicines outperforming a placebo, is inadequate to meaningfully measure the impact of psilocybin. That other clinical trials cannot determine differences in perception between psilocybin and LSD (mentioned above) is curious, as I've heard the difference is comparable to a chamois and steel wool. After >50 years of research the medical community have agreed that set and setting make a difference to a psilocybin experience. Thank you to all you advocates of safe practice; in the last two years I have been contacted by many people desperate to try psilocybin and improve their quality of life, and these people in long term pain will ultimately benefit from your knowledge. Shame about the whole 'going to jail for a mushroom' thing in the meantime. For those who want to research magic mushrooms It's possible the last two years were a bit of a failure academically. I received 0/6 grants (=up to six months of work writing grants and some delightfully harsh personal feedback), published one paper, another is in review here, and another is in preparation. This information translates to: there is ample room to find new knowledge, but you'll need a couple of planets to align if you want funding. I did this research on a Fellowship from covid stimulus for one year, and have worked part-time ever since. BioPlatforms Australia kicked in ~$17,000 (in-kind) for sequencing genomes, the rest were sequenced on the back of plant pathology funding leftovers. You need mentors who will give you freedom. I was told by nearly all my mentors not to work on magic mushrooms and that it would end my career. Well, my research career is probably ending, but if anything, magic mushrooms gave it more life and this has been the most interesting, exciting, and motivating topic in 16 years studying Fungi. Thank you to Kelly from BioPlatforms, Entheogenesis for helping share my journey, Louise, Tim James (for fresh ideas when sharing knowledge on Psilocybe with the scientific community), all of you who have sent me spores including, Snu, Jan T, Dave H, Sarah L, Matt W, Andy B, Rhys L, Tim S, Bock and Dan, Feryl Beryl Bunyip, Bill, Davie GGG, Josh B, Chris Apples, Chris M, Ema, Sequoia, Ronny, Greta Puls, and Caine :) I might make another one or two posts when the last work is published and to update on my next career stage... until then take it easy. Tchao for meow and thanks for reading. Alistair Taxonomists describe new species of fungi based on phenotypic differences reflected in morphology and ecology, and genetic differences, usually by comparison of conserved genes (like the ITS and LSU regions of ribosomal DNA). The challenge for taxonomists is to find a happy medium between lumping and splitting species. Species boundaries that are too broad cannot distinguish among actual differences (such as pathogenicity and geographic range), and lose meaning when too narrow and many names are applied across intraspecific variation. The ITS region is a biological gem. It can be amplified with universal primers across the fungal kingdom, it is interspecifically variable (usually), and intraspecifically conserved (usually). I fell in love with the ITS region working on smut fungi and rust fungi, both of which are more easily amplified with ITS primers than any other genes. But the ITS is not a Cinderella fit for all Fungi, there is not enough variation among species of Ascomycota (e.g., the ITS is not variable enough to distinguish species of Fusarium), and some familiarity is needed to understand the limitations of the ITS region in different groups. The Psilocybe subaeruginosa species complex has grown based on variation in the ITS region, phenotypic variability, and geographic range. It includes P. cyanescens, P. australiana, P. eucalypta, P. tasmaniana, P. azurescens, P. makarorae, P. weraroa, and P. allenii, the latter two described on differences in rDNA. Below is a phylogenetic hypothesis based on an alignment of the ITS region including all of the 86 sequenced genomes of P. subaeruginosa from Australia, and sequences taken from GenBank of other taxa in the complex. The ITS region is intraspecifically variable across the complex in Australia. This variation makes P. cyanescens, P. azurescens, and P. allenii paraphyletic in regard to P. subaeruginosa. A taxonomic solution would be to either call everything P. subaeruginosa, or potentially describe more and more species to reflect groupings. This 'splitting' option is unsatisfactory because it would not match the population analyses posted previously (essentially the relationships among ITS do not reflect population networks based on >1.5 million SNPs). My opinion is that P. subaeruginosa is one phenotypically variable species with a widespread distribution caused by saprotrophic invasions in the northern hemisphere. Describing more species in the complex ultimately adds more future synonyms.
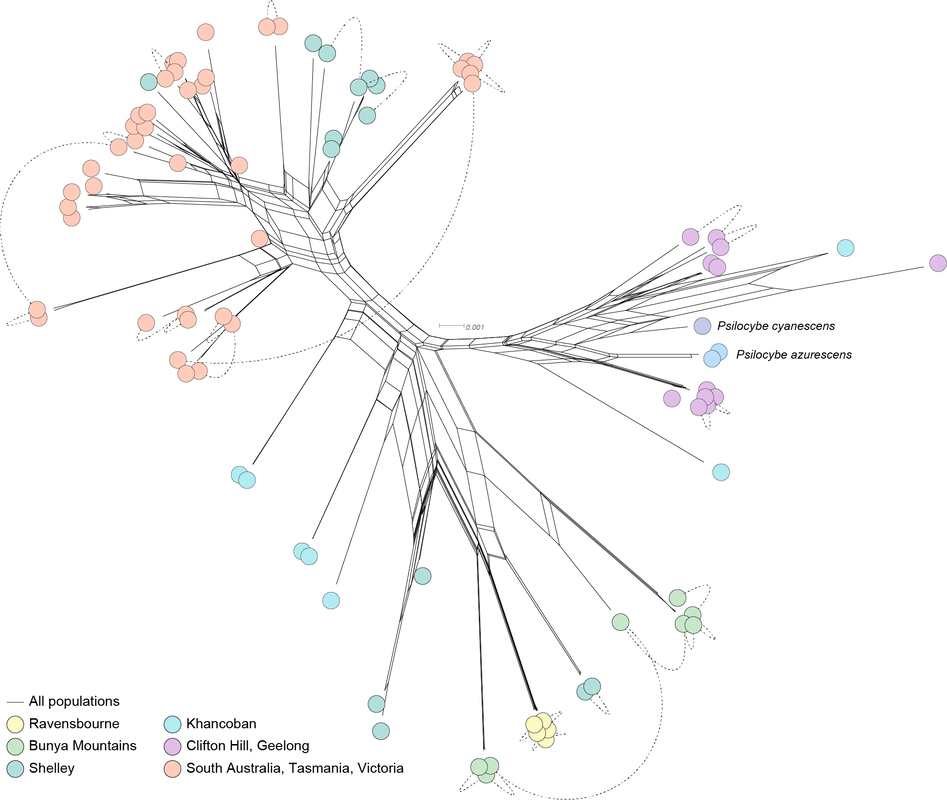
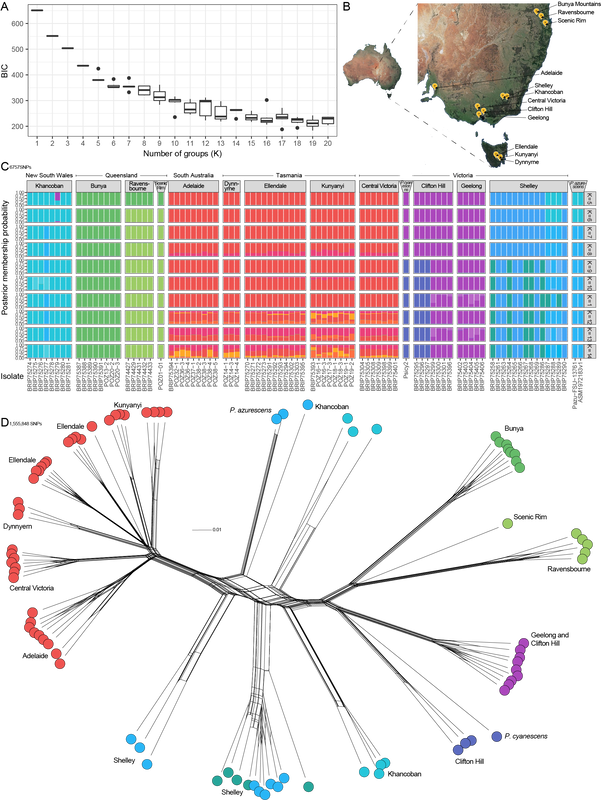 A. K-means test for number of groups in the data. The number of populations can be estimated from the 'elbow' in the graph. B. Sampling of P. subaeruginosa from Australia. C. DAPC analysis with facets designated by the final cutoff for K. Each horizontal row shows how the overall dataset changes if a population is split from the rest (new colours in each row are the 'new' population). D. SplitsTree network of relationships among 1.5 million SNPs in the Australia population of P. subaeruginosa, with three genomes of wood lovers from the Northern Hemisphere. Several of these populations include sibling genotypes (specifically Ravensbourne, Kunyanyi, Ellendale, Dynnyern, Clifton Hill). Another sub season is soon upon us. The story of how populations of P. subaeruginosa are structured in Australia is quite clear, even though we're missing samples from the entire length of NSW. Populations are structured by geography, with some localised, mountain-top populations in Queensland having few opportunities for outcrossing. Populations in southern Australia are divided by a mountain range. There is evidence of distribution of P. subaeruginosa in mulch, for example, the populations from Clifton Hill and Geelong (both mulched garden beds).
It's been fantastic to speak with the collectors of these mushrooms. Based on their information, it is likely mycelium of P. subaeruginosa is persistent from year to year. This persistence keeps the same genotypes at the same locations, which is different to P. cubensis for which genotypes are ephemeral and dependent on spore distribution in manure (in my opinion). A comparable example to P. subaeruginosa is the humungous fungus among us, which is a huge clonal network of mycelium. Mycelia of P. subaeruginosa is ubiquitous in natural Australian environments, and those of you who re-visit mushrooms from season-to-season see the same genotype every time. There are opportunities for outcrossing, especially with mulch distribution and artificial movement of soils and wood chips. Psilocybe azurescens and P. cyanescens are introduced to the northern hemisphere from Australia. They cluster nicely among our populations from Australia, although we weren't lucky enough to sample their exact provenance in this study. For context, much of the sampling is of siblings, sampled from sister basidiospores from single mushroom caps. Keep in mind that many of the genomes in this figure are offspring and that will put the genetic distance in context. The genome of P. subaeruginosa is about 50 million base pairs, and I'm looking at a tiny fraction of differences (1.5 million base pairs) to illustrate how these genomes are related. I think these data support P. subaeruginosa is one species and other related taxa are synonyms. My opinion is no indication of the future taxonomy, and many taxonomists prefer to split groups of microorganisms into different species. There is a lot of variation at the psilocybin locus in our sampled populations of P. subaeruginosa. It's about to get interesting if we can link any of these variations to Wood Lover's Paralysis. Should have something for you in the next couple of weeks. The P. cubensis study is finished. It's sitting with my co-authors for their magic touch. Looking forward to sharing some new knowledge about mating in cubes with y'all as well :). Apologies for the delay. At last, the data are in a state to analyse. We ended up with 85 genomes funded by BioPlatforms Australia, of which 80 were Psilocybe subaeruginosa.
The data are assembled and annotated, and the below figure is a way to visualise the relationships among the sampled populations, plus three genomes from GenBank of P. azurescens and P. cyanescens. The most obvious take home at this early stage is P. azurescens and P. cyanescens are likely conspecific with P. subaeruginosa. No surprises here, this was always the hypothesis. But good to see it play out the way we expected. These data aren't perfect yet and I'm still finalising the SNPs. More information coming, especially on the psilocybin gene cluster, soon! Most of my time at the moment is toward publishing the analyses of P. cubensis and the story behind their introduction to Australia. We've resolved mating in cubes, which is similar to P. subaeruginosa, but with more red herrings for compatibility. I'll tease this out a tad more soon. @Feral Beryl Bunyip, I sequenced a genome of one of the mushrooms you sent me. Give me a shout if you have dreams of what should happen with this treat :). I've been contacted by many Australians interested in volunteering for clinical trials. The jam is I work on the evolution of mushrooms and don't run clinical trials.
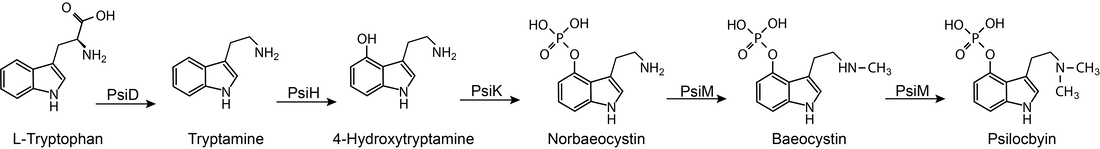
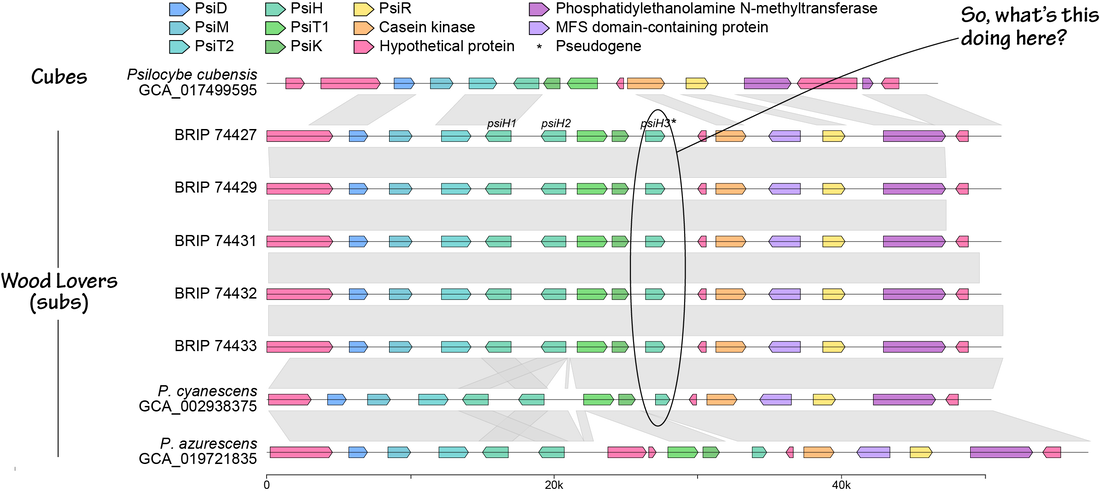
Here's an opportunity for those of you who microdose psilocybin and live in Sydney, Australia. Dr Vince Polito in the School of Psychological Sciences and his team at Macquarie University are looking for individuals who microdose (or are already planning to microdose) using psilocybin or ‘magic mushrooms’. Participants will visit their lab on two occasions: once after taking a microdose and once after taking a placebo. They will use an MEG scanner to explore what happens in peoples’ brains when they microdose. Added bonus, they can reward your participation financially! For more information and to sign up, please visit microdosingstudy.com or email [email protected] You can find more about Vince Polito’s research here. You can read more about their study here. Thank you Ronny, Eva, Jay, Jacob, Nicole, Billy, Sophie, and Caine for taking care of us and all the hospitality at Entheogenesis. Fun on a bun, and fantastic to meet so many of the people who have sent me spores this year. After my talk, which you can watch here if you attended the conference, I spoke to more than 10 people about their experiences of Wood Lover's Paralysis, which occurs as a side effect when eating Psilocybe subaeruginosa, and temporarily paralyses people during a psilocybin experience. The grey areas around the cause of WLP are: (i) is there variation among humans (are there more susceptible people), (ii) is it caused by metabolism when mushrooms grow on particular substrates, or (iii) is it caused by secondary metabolites produced by genotypes of P. subaeruginosa? It felt like the majority of you have experienced WLP and with varying symptoms (from full paralysis including not being able to speak, to paralysis of limbs). We might be able to rule out that some people are immune, even though one member of our community has never experienced WLP after >30 years of practice. The anecdotal evidence leans toward substrate or genotype. Paralysis occurred in people who had picked from wild areas and cultivated garden beds, and paralysis reoccurred when people ate smaller doses of the same batch of harvested mushrooms (interesting). In a manuscript coming out soon (it's been reviewed, just stuck in a bottleneck), we show an additional copy of one of the genes that produce an enzyme in the metabolic pathway of psilocybin. PsiH genes convert tryptamine into 4- Hydroxytryptamine, but when you have redundant copies of a gene, they can start to innovate. Redundant genes are not under selection (their previous function is wrapped up by the original copy) and they can explore mutations over generations to take on versatile functions. If the new enzymes produced are not deleterious and confer a slight advantage, they'll stick around in the gene pool.
The first hypothesis to explore is that there are functional copies of the psiH3 gene in the population of P. subaeruginosa, and individuals with those copies produce a Wood Lover's Paralysis phenotype. Those data should be coming to me soon (it is Christmas after all) and we can all share in the excitement. |
Designer Shrooms @ Funky Fungus on 1st July 2023
I started a gig at Funky Fungus as Chief Scientific Officer to make designer shrooms Our research on Psilocybe
|